Genetic Lab Testing with Consultation
3X4 Genetics Blueprint + Eileen Durfee Consultation
If it impacts your health, it belongs in your plan. The 3X4 Genetics Blueprint translates key genetic variants into clear, pathway-based health insights—covering 36 metabolic pathways across 6 health categories—so you can focus on the areas that matter most first.
Your genetics don’t dictate your destiny—but they can reveal where you’re more sensitive and what to prioritize. This report is designed to turn complex genetic data into a smarter, more personal plan for nutrition, lifestyle, training, and recovery.
When you order through Eileen Durfee, you get more than a test—you get support around the whole process so you feel confident from purchase to plan:
- Help choosing the right testing option for your goals (so you don’t overbuy or under-test)
- Guidance on what to expect, how to use the report, and how to turn results into priorities
- A streamlined path to consultation scheduling and next-step planning
- A practitioner’s lens focused on action: food, lifestyle, training, recovery, and realistic sequencing
What you receive
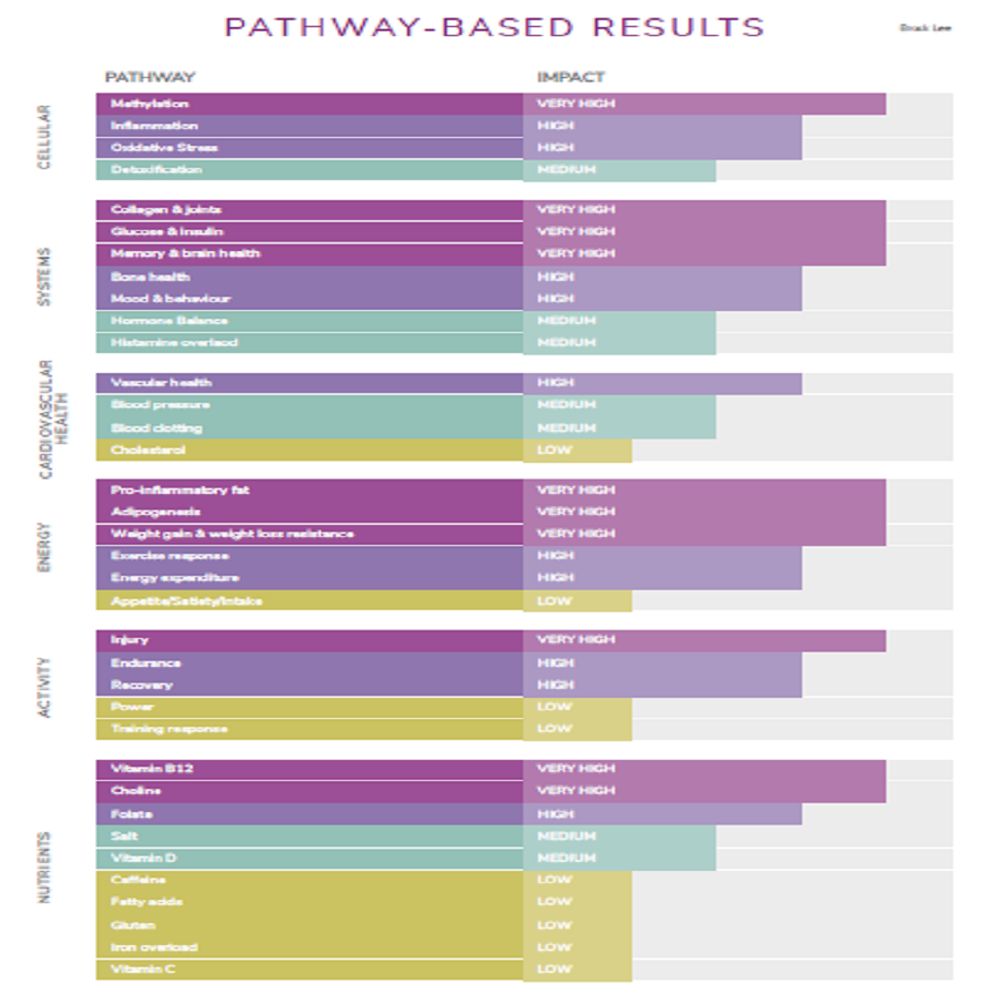
- Your pathway impact dashboard (from highest to lowest impact using easy color-coding) so priorities are obvious.
- Gene results organized by pathway, not dumped as raw data—so patterns appear (inflammation + detox + glucose control, etc.).
- A framework that connects your results to practical levers: nutrition, lifestyle, training, and recovery.
Why Eileen Durfee’s consultation makes it better
Many genetic reports are informative—but implementation is where people stall. The Blueprint itself emphasizes that the most useful insights emerge when genetic findings are combined with your history, diet, lifestyle, and goals in the hands of an expert practitioner.
With Eileen, you’re not just “reviewing genes.” You’re turning them into a personalized action plan, including:
- What to address first (highest-impact pathways)
- What’s likely driving your symptoms or plateaus (cross-pathway patterns)
- How to tailor food, supplements, stress, sleep, and training to your genetics
- Clear next steps you can actually follow
Why this approach is superior to typical testing
- Pathway-level interpretation (systems thinking): you see how genes interact inside real biology, not isolated SNP trivia.
- Prioritization built in: impact scoring + color-coding prevents overwhelm and guesswork.
- Practitioner-guided personalization: your results become a strategy, not a PDF that sits in a folder.
How many genes are tested?
137 genes/markers are included.
How these genes connect to pathways in 3X4 Genetics
In the 3X4 Genetics Blueprint, gene variants are organized into pathway-based results (so you’re not left with a disconnected SNP list). The report covers 36 metabolic pathways across 6 health categories, and it calculates an impact level for each pathway using color-coding to show where to focus first.
Below is a practical “gene → pathway” map (the same gene can influence more than one pathway).
Gene explanations by pathway
1) Cellular pathways
A) Methylation (methyl donors, one-carbon cycle, neurotransmitter balance)
| Gene(s) | Explanation |
|---|---|
| MTHFR | folate activation (methyl cycle throughput) |
| MTHFD1 | folate cycle support (one-carbon metabolism) |
| MTR | remethylates homocysteine to methionine |
| MTRR | regenerates active B12 for MTR |
| BHMT | alternate homocysteine “shortcut” using betaine/choline |
| CBS | diverts homocysteine toward transsulfuration (sulfur pathway) |
| TCN2 | vitamin B12 transport into cells |
| PEMT | endogenous choline/phosphatidylcholine production |
| CHDH | choline → betaine (feeds BHMT methylation) |
B) Inflammation (cytokines, acute phase response, immune signaling)
| Gene(s) | Explanation |
|---|---|
| TNFA | inflammatory signaling intensity |
| IL-1 | inflammatory activation signaling (marker) |
| IL-6 / IL-6R | cytokine signaling and receptor sensitivity |
| CRP | downstream inflammation/acute phase marker |
| HLA | immune recognition; links to gluten/autoimmune susceptibility |
| PPARA / PPARG / PPARD | metabolic inflammation switches (fat/glucose crosstalk) |
| SIRT1 | inflammation control via energy-sensing pathways |
| FOXO1 / FOXO3 | stress-response transcription factors |
C) Oxidative stress (antioxidant enzymes, mitochondrial protection)
| Gene(s) | Explanation |
|---|---|
| CAT | breaks down hydrogen peroxide |
| MNSOD (SOD2) | mitochondrial superoxide defense |
| GPX1 | peroxide neutralization (glutathione system) |
| NQO1 | quinone detox / antioxidant defense |
| NRF2 | master “turn on antioxidant genes” regulator |
| OGG1 | oxidative DNA repair |
| UCP1 / UCP2 / UCP3 | mitochondrial uncoupling; ROS/energy balance |
D) Detoxification (Phase I/II detox, clearance, xenobiotics)
| Gene(s) | Explanation |
|---|---|
| Phase I (activation/processing) | |
| CYP1A1 / CYP1A2 / CYP1B1 / CYP2C19 / CYP2C9 / CYP2D6 / CYP3A4 | drug/hormone/toxin metabolism |
| EPHX1 | epoxide detox (often paired with CYP activity) |
| ALDH2 | aldehyde clearance (alcohol and oxidative byproducts) |
| Phase II (conjugation/clearance) | |
| GSTM1 / GSTT1 / GSTP1 / GSTO2 | glutathione conjugation capacity |
| NAT1 / NAT2 | acetylation (amine/chemical handling) |
| SULT1A1 | sulfation (hormones/phenols) |
| UGT2B15 / UGT2B17 | glucuronidation (hormones/xenobiotics) |
| PON1 | antioxidant/organophosphate handling (cardio + detox overlap) |
2) Systems pathways
A) Collagen & joints (connective tissue structure + breakdown/repair)
| Gene(s) | Explanation |
|---|---|
| COL1A1 / COL3A1 / COL12A1 | collagen structural proteins |
| GDF5 | joint/cartilage development and resilience |
| MMP1 / MMP2 / MMP3 | collagen breakdown/remodeling enzymes |
| TIMP4 | MMP inhibition (controls breakdown) |
| VEGFA | tissue perfusion/healing support (also vascular) |
B) Glucose & insulin (glycemic regulation, insulin signaling, storage)
| Gene(s) | Explanation |
|---|---|
| TCF7L2 | insulin/glucose regulation risk signaling |
| IRS1 | insulin receptor signaling efficiency |
| SLC2A2 | glucose transport (GLUT2) |
| PPARG / PPARGC1A / PPARA / PPARD | insulin sensitivity + fuel switching |
| FTO | appetite/energy balance associations |
| MTNR1B | melatonin receptor; glucose timing/fasting glucose associations |
| ADIPOQ | adiponectin signaling; insulin sensitivity support |
| LEPR | leptin signaling; appetite + metabolic control |
C) Memory & brain health (neuroplasticity, vascular brain support, aging risk)
| Gene(s) | Explanation |
|---|---|
| APOE | lipid transport in brain; cognitive aging associations |
| TOMM40 | mitochondrial import; cognition/aging associations |
| BDNF | neuroplasticity/learning; stress response |
| COMT | dopamine breakdown; cognition/stress tolerance overlap |
| CACNA1C | neuronal calcium signaling |
| AKT1 | neuronal survival/signaling pathways |
| NOS3/ENOS | blood flow support (brain perfusion overlap) |
D) Bone health (vitamin D axis + collagen structure)
| Gene(s) | Explanation |
|---|---|
| VDR | vitamin D receptor function |
| GC | vitamin D binding/transport |
| CYP2R1 | vitamin D activation step |
| COL1A1 | bone matrix collagen backbone |
E) Mood & behaviour (serotonin, dopamine, GABA, reward, bonding)
| Gene(s) | Explanation |
|---|---|
| Serotonin signaling | |
| 5HT2A | serotonin receptor signaling (mood, sleep, perception) |
| HTR1A | serotonin receptor regulation (stress/mood) |
| Dopamine/reward | |
| DRD1 / DRD3 / DRD4 | dopamine receptors (motivation/novelty/reward) |
| ANKK1/DRD2 | dopamine D2 pathway marker (reward/addiction tendency) |
| Breakdown enzymes / neurotransmitter balance | |
| MAOA | monoamine breakdown (serotonin/dopamine/norepinephrine) |
| COMT | dopamine/norepinephrine breakdown (stress/cognition) |
| GAD1 | makes GABA (calming neurotransmitter) |
| GABRA2 | GABA receptor signaling (calm/anxiety traits) |
| Reward, stress buffering, connection | |
| OPRM1 | opioid receptor (reward/pain response) |
| FAAH | endocannabinoid breakdown (stress, reward, inflammation overlap) |
| OXTR | oxytocin receptor (bonding/social stress resilience) |
| ANK3 / CACNA1C | neuronal excitability/mood stability markers |
| CHRNA5 | nicotinic receptor (stimulation/addiction tendency) |
F) Hormone balance (estrogen/androgen metabolism + binding/clearance)
| Gene(s) | Explanation |
|---|---|
| CYP17A1 | sex steroid synthesis step |
| CYP19A1 | aromatase (androgen → estrogen) |
| ESR2 | estrogen receptor beta signaling |
| SHBG | hormone binding/availability |
| SRD5A1 | androgen conversion (DHT pathway) |
| UGT2B15 / UGT2B17 / SULT1A1 | hormone clearance (phase II) |
G) Histamine overload (breakdown capacity)
| Gene(s) | Explanation |
|---|---|
| DAO | histamine breakdown in gut/periphery |
| HNMT | histamine breakdown inside cells |
| (Also influenced by inflammation genes like IL-6/TNFA) | |
3) Cardiovascular health pathways
A) Vascular health (endothelial function, nitric oxide, repair)
| Gene(s) | Explanation |
|---|---|
| NOS3 / ENOS | nitric oxide production; vessel flexibility |
| VEGF / VEGFA / VEGFR2 | vascular growth/repair signaling |
| HO-1 (HMOX1) | oxidative/inflammatory vessel protection |
| CRP / OGG1 / NRF2 | inflammation/oxidative stress impacts vessels |
B) Blood pressure (RAAS signaling + adrenergic tone)
| Gene(s) | Explanation |
|---|---|
| ACE / ACE2 | angiotensin processing balance |
| AGT | angiotensinogen supply |
| AGTR1 / AGTR2 | angiotensin receptor signaling |
| REN | renin initiation of RAAS cascade |
| ADRB2 | adrenergic tone (stress response, vascular constriction/dilation) |
C) Blood clotting (coagulation risk tendency)
| Gene(s) | Explanation |
|---|---|
| F2 | prothrombin pathway |
| F5 | Factor V clotting regulation |
| HPA-1 | platelet adhesion marker |
D) Cholesterol & lipids (transport, triglycerides, HDL dynamics)
| Gene(s) | Explanation |
|---|---|
| APOE | lipoprotein transport/clearance |
| APOA5 / APOC3 | triglyceride handling |
| APOA2 | fat response signaling (diet-fat sensitivity traits) |
| CETP | HDL/LDL particle exchange |
| LPL | triglyceride breakdown from particles |
| FABP2 | intestinal fat absorption/handling |
| PON1 | HDL antioxidant function |
4) Energy pathways (fat storage, release, appetite, metabolism)
A) Pro-inflammatory fat
| Gene(s) | Explanation |
|---|---|
| TNFA / IL-1 / IL-6 / IL-6R / CRP | adipose inflammation signaling |
| ADIPOQ | anti-inflammatory adipokine support |
| PPARG / PPARA / SIRT1 | inflammation ↔ metabolism switches |
B) Adipogenesis (fat-cell creation/storage propensity)
| Gene(s) | Explanation |
|---|---|
| PPARG | adipocyte differentiation “master switch” |
| PLIN | fat droplet storage dynamics |
| ADRB3 / ADRB2 | fat mobilization signaling |
| LEPR | leptin feedback (storage vs burn) |
| MMP2 | tissue remodeling that can affect adipose expansion |
C) Weight gain & weight loss resistance
| Gene(s) | Explanation |
|---|---|
| FTO | weight regulation associations |
| MC4R | appetite/satiety control center signaling |
| CLOCK | circadian rhythm ↔ metabolism coupling |
| UCP1/2/3 | energy expenditure/thermogenesis tendency |
| TCF7L2 / IRS1 | glucose signaling that affects storage patterns |
D) Exercise response & energy expenditure
| Gene(s) | Explanation |
|---|---|
| PPARGC1A / PPARD / PPARA | mitochondrial biogenesis & fuel switching |
| ADRB2 / ADRB3 | fat mobilization during activity |
| UCP genes | resting burn/thermogenesis traits |
E) Appetite / satiety / intake
| Gene(s) | Explanation |
|---|---|
| LEPR / MC4R / FTO | hunger/fullness signaling balance |
| TAS2R38 | bitter taste sensitivity (food preference effects) |
5) Activity pathways (training, performance, injury risk)
A) Injury / connective tissue recovery
| Gene(s) | Explanation |
|---|---|
| COL1A1 / COL3A1 / COL12A1 / GDF5 | tissue strength |
| MMP3 / TIMP4 | remodeling balance |
| VEGFA | healing blood supply support |
| TNFA / CRP | inflammation that can impair recovery |
B) Endurance / oxygen utilization
| Gene(s) | Explanation |
|---|---|
| ACE | endurance vs power tendency marker |
| HIF1A | oxygen response signaling |
| NOS3 | blood flow support |
| VEGF | capillary growth signaling |
| PPARGC1A / PPARA | aerobic capacity support pathways |
C) Power / sprint traits
| Gene(s) | Explanation |
|---|---|
| ACTN3 | fast-twitch muscle performance marker |
| CKM | muscle energy buffering |
| AMPD1 | ATP recycling during intense bursts |
D) Training response (adaptation rate)
| Gene(s) | Explanation |
|---|---|
| ACE / ACTN3 / PPARGC1A / NRF2 | adaptation, recovery, oxidative load handling |
| ACSL1 | fatty acid activation for energy use in muscle |
6) Nutrients pathways (absorption, activation, utilization)
A) Vitamin B12
| Gene(s) | Explanation |
|---|---|
| TCN2 | transport into cells |
| FUT2 | gut/secretor status; influences B12 availability via microbiome dynamics |
B) Choline
| Gene(s) | Explanation |
|---|---|
| PEMT / CHDH / BHMT | choline need & use in methylation/membranes |
C) Folate
| Gene(s) | Explanation |
|---|---|
| MTHFR / MTHFD1 | folate processing and availability |
D) Salt sensitivity
| Gene(s) | Explanation |
|---|---|
| ACE / AGT / AGTR1 / REN | RAAS control of fluid and pressure |
E) Vitamin D
| Gene(s) | Explanation |
|---|---|
| VDR | receptor response |
| GC | transport protein |
| CYP2R1 | activation step |
F) Caffeine
| Gene(s) | Explanation |
|---|---|
| CYP1A2 | primary caffeine metabolism speed |
| COMT | stimulant sensitivity via catechol handling |
G) Fatty acids
| Gene(s) | Explanation |
|---|---|
| FADS1 / FADS2 | omega-3/omega-6 conversion efficiency |
| ELOVL2 | long-chain fatty acid elongation |
| APOA2 / APOA5 / FABP2 | dietary fat response/handling |
H) Gluten
| Gene(s) | Explanation |
|---|---|
| HLA | celiac-associated immune recognition markers |
I) Iron overload
| Gene(s) | Explanation |
|---|---|
| HFE | iron absorption regulation |
J) Vitamin C
| Gene(s) | Explanation |
|---|---|
| SLC23A1 | vitamin C transport into cells |
FAQ — Genetic Lab Testing with Consultation (3X4 Genetics Blueprint)
1. What is this product?
It’s Genetic Lab Testing with Consultation, built around the 3X4 Genetics Blueprint, which translates key genetic variants into pathway-based health insights across 36 metabolic pathways in 6 health categories.
2. What’s included with the purchase?
You receive a pathway impact dashboard ranked from highest to lowest impact with easy color-coding, gene results organized by pathway (instead of raw data) to help patterns show up, and a framework connecting results to practical levers like nutrition, lifestyle, training, and recovery.
3. What will Eileen help you do during the consultation?
The consultation is focused on turning gene insights into a personalized action plan, including what to address first (highest-impact pathways), what they are driving symptoms or plateaus (cross-pathway patterns), how to tailor food, supplements, stress, sleep, and training to your genetics, and clear, followable next steps.
4. How is this different from “typical” genetic testing reports?
This approach emphasizes pathway-level interpretation (systems thinking) rather than isolated SNPs, built-in prioritization via impact scoring and color-coding, and practitioner-guided personalization so results become a strategy (not just a report).
5. How many genes/markers are included?
The page states 137 genes/markers are included.
6. How are results organized in the report?
Gene variants are organized into pathway-based results, and an impact level is calculated for each pathway using color-coding to show where to focus first.
7. What are the main health categories/pathway groups covered?
The report covers 36 metabolic pathways across 6 health categories, organized into pathway groups like Cellular pathways, Systems pathways, Cardiovascular health pathways, Energy pathways, Activity pathways, and Nutrients pathways.
8. What “cellular pathways” are included?
Methylation, inflammation, oxidative stress, and detoxification (Phase I & Phase II).
9. What “systems pathways” are included?
Collagen & joints, glucose & insulin, memory & brain health, bone health, mood & behaviour, hormone balance, and histamine overload.
10. What cardiovascular pathways are included?
Vascular health, blood pressure, blood clotting, and cholesterol & lipids.
11. What energy-related pathways are included?
Pro-inflammatory fat, adipogenesis, weight gain & weight loss resistance, exercise response & energy expenditure, and appetite / satiety / intake.
12. What activity/training pathways are included?
Injury / connective tissue recovery, endurance / oxygen utilization, power / sprint traits, and training response (adaptation rate).
13. What nutrient-related pathways are included?
Vitamin B12, Choline, Folate, Salt sensitivity, Vitamin D, Caffeine, Fatty acids, Gluten, Iron overload, and Vitamin C.