Genetic Lab Testing with Consultation 2
Genetic Lab Testing with Consultation
Your DNA is information. This service turns it into a plan.
Genetic Lab Testing with Consultation analyzes 137 key genes and organizes your results into the biological pathways that drive real-world outcomes—inflammation, detoxification, glucose/insulin control, cardiovascular function, neurotransmitter balance (mood & cravings), nutrient needs, and performance/recovery.
Then you meet 1:1 with Eileen Durfee to translate the report into a clear, personalized strategy—so you know what to focus on first, what to change, and what to ignore.
What makes this different
Most genetic tests give you a long list of SNPs and generic advice. This program is built for action:
- Pathway-based interpretation (how genes work together—not isolated trivia)
- Clear prioritization so you’re not overwhelmed
- Expert consultation to turn findings into practical next steps
What you’ll walk away with
- A smarter nutrition approach based on your genetics (carb/fat response, appetite signaling, and timing)
- Clarity on inflammation patterns and recovery speed
- Insight into detox and oxidative stress capacity (how you handle caffeine, alcohol, hormones, and environmental load)
- Nutrient utilization patterns (methylation, vitamin D, choline/B12/folate pathways, and more)
- Performance and injury-resilience tendencies (endurance vs power traits, tissue remodeling, recovery needs)
What’s included
- At-home genetic collection kit
- 137-gene pathway-based report
- 1:1 consultation with Eileen Durfee
- Personalized action roadmap (nutrition, lifestyle, supplements, training, recovery)
Best for you if you feel…
- Stuck despite “doing everything right”
- Confused by conflicting diet/supplement advice
- Slower recovery, stubborn weight, fatigue, brain fog, cravings, or inflammation
- Ready for a plan that’s tailored to your biology
Order Genetic Lab Testing with Consultation to stop guessing—and start making decisions that match your genetics.
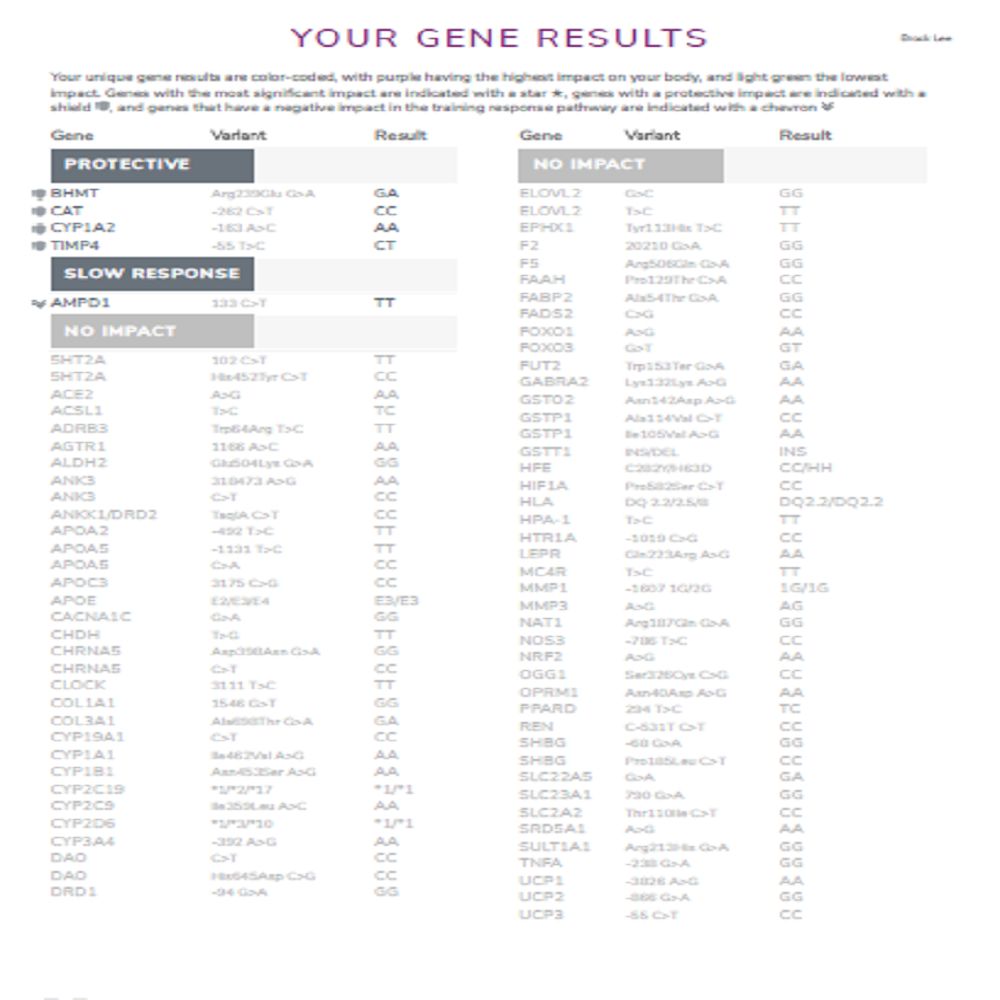
How many genes are tested?
137 genes/markers are included.
How these genes connect to pathways in 3X4 Genetics
In the 3X4 Genetics Blueprint, gene variants are organized into pathway-based results (so you’re not left with a disconnected SNP list). The report covers 36 metabolic pathways across 6 health categories, and it calculates an impact level for each pathway using color-coding to show where to focus first.
Below is a practical “gene → pathway” map (the same gene can influence more than one pathway).
Gene explanations by pathway
1) Cellular pathways
A) Methylation (methyl donors, one-carbon cycle, neurotransmitter balance)
| Gene(s) | Explanation |
|---|---|
| MTHFR | folate activation (methyl cycle throughput) |
| MTHFD1 | folate cycle support (one-carbon metabolism) |
| MTR | remethylates homocysteine to methionine |
| MTRR | regenerates active B12 for MTR |
| BHMT | alternate homocysteine “shortcut” using betaine/choline |
| CBS | diverts homocysteine toward transsulfuration (sulfur pathway) |
| TCN2 | vitamin B12 transport into cells |
| PEMT | endogenous choline/phosphatidylcholine production |
| CHDH | choline → betaine (feeds BHMT methylation) |
B) Inflammation (cytokines, acute phase response, immune signaling)
| Gene(s) | Explanation |
|---|---|
| TNFA | inflammatory signaling intensity |
| IL-1 | inflammatory activation signaling (marker) |
| IL-6 / IL-6R | cytokine signaling and receptor sensitivity |
| CRP | downstream inflammation/acute phase marker |
| HLA | immune recognition; links to gluten/autoimmune susceptibility |
| PPARA / PPARG / PPARD | metabolic inflammation switches (fat/glucose crosstalk) |
| SIRT1 | inflammation control via energy-sensing pathways |
| FOXO1 / FOXO3 | stress-response transcription factors |
C) Oxidative stress (antioxidant enzymes, mitochondrial protection)
| Gene(s) | Explanation |
|---|---|
| CAT | breaks down hydrogen peroxide |
| MNSOD (SOD2) | mitochondrial superoxide defense |
| GPX1 | peroxide neutralization (glutathione system) |
| NQO1 | quinone detox / antioxidant defense |
| NRF2 | master “turn on antioxidant genes” regulator |
| OGG1 | oxidative DNA repair |
| UCP1 / UCP2 / UCP3 | mitochondrial uncoupling; ROS/energy balance |
D) Detoxification (Phase I/II detox, clearance, xenobiotics)
| Gene(s) | Explanation |
|---|---|
| Phase I (activation/processing) | |
| CYP1A1 / CYP1A2 / CYP1B1 / CYP2C19 / CYP2C9 / CYP2D6 / CYP3A4 | drug/hormone/toxin metabolism |
| EPHX1 | epoxide detox (often paired with CYP activity) |
| ALDH2 | aldehyde clearance (alcohol and oxidative byproducts) |
| Phase II (conjugation/clearance) | |
| GSTM1 / GSTT1 / GSTP1 / GSTO2 | glutathione conjugation capacity |
| NAT1 / NAT2 | acetylation (amine/chemical handling) |
| SULT1A1 | sulfation (hormones/phenols) |
| UGT2B15 / UGT2B17 | glucuronidation (hormones/xenobiotics) |
| PON1 | antioxidant/organophosphate handling (cardio + detox overlap) |
2) Systems pathways
A) Collagen & joints (connective tissue structure + breakdown/repair)
| Gene(s) | Explanation |
|---|---|
| COL1A1 / COL3A1 / COL12A1 | collagen structural proteins |
| GDF5 | joint/cartilage development and resilience |
| MMP1 / MMP2 / MMP3 | collagen breakdown/remodeling enzymes |
| TIMP4 | MMP inhibition (controls breakdown) |
| VEGFA | tissue perfusion/healing support (also vascular) |
B) Glucose & insulin (glycemic regulation, insulin signaling, storage)
| Gene(s) | Explanation |
|---|---|
| TCF7L2 | insulin/glucose regulation risk signaling |
| IRS1 | insulin receptor signaling efficiency |
| SLC2A2 | glucose transport (GLUT2) |
| PPARG / PPARGC1A / PPARA / PPARD | insulin sensitivity + fuel switching |
| FTO | appetite/energy balance associations |
| MTNR1B | melatonin receptor; glucose timing/fasting glucose associations |
| ADIPOQ | adiponectin signaling; insulin sensitivity support |
| LEPR | leptin signaling; appetite + metabolic control |
C) Memory & brain health (neuroplasticity, vascular brain support, aging risk)
| Gene(s) | Explanation |
|---|---|
| APOE | lipid transport in brain; cognitive aging associations |
| TOMM40 | mitochondrial import; cognition/aging associations |
| BDNF | neuroplasticity/learning; stress response |
| COMT | dopamine breakdown; cognition/stress tolerance overlap |
| CACNA1C | neuronal calcium signaling |
| AKT1 | neuronal survival/signaling pathways |
| NOS3/ENOS | blood flow support (brain perfusion overlap) |
D) Bone health (vitamin D axis + collagen structure)
| Gene(s) | Explanation |
|---|---|
| VDR | vitamin D receptor function |
| GC | vitamin D binding/transport |
| CYP2R1 | vitamin D activation step |
| COL1A1 | bone matrix collagen backbone |
E) Mood & behaviour (serotonin, dopamine, GABA, reward, bonding)
| Gene(s) | Explanation |
|---|---|
| Serotonin signaling | |
| 5HT2A | serotonin receptor signaling (mood, sleep, perception) |
| HTR1A | serotonin receptor regulation (stress/mood) |
| Dopamine/reward | |
| DRD1 / DRD3 / DRD4 | dopamine receptors (motivation/novelty/reward) |
| ANKK1/DRD2 | dopamine D2 pathway marker (reward/addiction tendency) |
| Breakdown enzymes / neurotransmitter balance | |
| MAOA | monoamine breakdown (serotonin/dopamine/norepinephrine) |
| COMT | dopamine/norepinephrine breakdown (stress/cognition) |
| GAD1 | makes GABA (calming neurotransmitter) |
| GABRA2 | GABA receptor signaling (calm/anxiety traits) |
| Reward, stress buffering, connection | |
| OPRM1 | opioid receptor (reward/pain response) |
| FAAH | endocannabinoid breakdown (stress, reward, inflammation overlap) |
| OXTR | oxytocin receptor (bonding/social stress resilience) |
| ANK3 / CACNA1C | neuronal excitability/mood stability markers |
| CHRNA5 | nicotinic receptor (stimulation/addiction tendency) |
F) Hormone balance (estrogen/androgen metabolism + binding/clearance)
| Gene(s) | Explanation |
|---|---|
| CYP17A1 | sex steroid synthesis step |
| CYP19A1 | aromatase (androgen → estrogen) |
| ESR2 | estrogen receptor beta signaling |
| SHBG | hormone binding/availability |
| SRD5A1 | androgen conversion (DHT pathway) |
| UGT2B15 / UGT2B17 / SULT1A1 | hormone clearance (phase II) |
G) Histamine overload (breakdown capacity)
| Gene(s) | Explanation |
|---|---|
| DAO | histamine breakdown in gut/periphery |
| HNMT | histamine breakdown inside cells |
| (Also influenced by inflammation genes like IL-6/TNFA) | |
3) Cardiovascular health pathways
A) Vascular health (endothelial function, nitric oxide, repair)
| Gene(s) | Explanation |
|---|---|
| NOS3 / ENOS | nitric oxide production; vessel flexibility |
| VEGF / VEGFA / VEGFR2 | vascular growth/repair signaling |
| HO-1 (HMOX1) | oxidative/inflammatory vessel protection |
| CRP / OGG1 / NRF2 | inflammation/oxidative stress impacts vessels |
B) Blood pressure (RAAS signaling + adrenergic tone)
| Gene(s) | Explanation |
|---|---|
| ACE / ACE2 | angiotensin processing balance |
| AGT | angiotensinogen supply |
| AGTR1 / AGTR2 | angiotensin receptor signaling |
| REN | renin initiation of RAAS cascade |
| ADRB2 | adrenergic tone (stress response, vascular constriction/dilation) |
C) Blood clotting (coagulation risk tendency)
| Gene(s) | Explanation |
|---|---|
| F2 | prothrombin pathway |
| F5 | Factor V clotting regulation |
| HPA-1 | platelet adhesion marker |
D) Cholesterol & lipids (transport, triglycerides, HDL dynamics)
| Gene(s) | Explanation |
|---|---|
| APOE | lipoprotein transport/clearance |
| APOA5 / APOC3 | triglyceride handling |
| APOA2 | fat response signaling (diet-fat sensitivity traits) |
| CETP | HDL/LDL particle exchange |
| LPL | triglyceride breakdown from particles |
| FABP2 | intestinal fat absorption/handling |
| PON1 | HDL antioxidant function |
4) Energy pathways (fat storage, release, appetite, metabolism)
A) Pro-inflammatory fat
| Gene(s) | Explanation |
|---|---|
| TNFA / IL-1 / IL-6 / IL-6R / CRP | adipose inflammation signaling |
| ADIPOQ | anti-inflammatory adipokine support |
| PPARG / PPARA / SIRT1 | inflammation ↔ metabolism switches |
B) Adipogenesis (fat-cell creation/storage propensity)
| Gene(s) | Explanation |
|---|---|
| PPARG | adipocyte differentiation “master switch” |
| PLIN | fat droplet storage dynamics |
| ADRB3 / ADRB2 | fat mobilization signaling |
| LEPR | leptin feedback (storage vs burn) |
| MMP2 | tissue remodeling that can affect adipose expansion |
C) Weight gain & weight loss resistance
| Gene(s) | Explanation |
|---|---|
| FTO | weight regulation associations |
| MC4R | appetite/satiety control center signaling |
| CLOCK | circadian rhythm ↔ metabolism coupling |
| UCP1/2/3 | energy expenditure/thermogenesis tendency |
| TCF7L2 / IRS1 | glucose signaling that affects storage patterns |
D) Exercise response & energy expenditure
| Gene(s) | Explanation |
|---|---|
| PPARGC1A / PPARD / PPARA | mitochondrial biogenesis & fuel switching |
| ADRB2 / ADRB3 | fat mobilization during activity |
| UCP genes | resting burn/thermogenesis traits |
E) Appetite / satiety / intake
| Gene(s) | Explanation |
|---|---|
| LEPR / MC4R / FTO | hunger/fullness signaling balance |
| TAS2R38 | bitter taste sensitivity (food preference effects) |
5) Activity pathways (training, performance, injury risk)
A) Injury / connective tissue recovery
| Gene(s) | Explanation |
|---|---|
| COL1A1 / COL3A1 / COL12A1 / GDF5 | tissue strength |
| MMP3 / TIMP4 | remodeling balance |
| VEGFA | healing blood supply support |
| TNFA / CRP | inflammation that can impair recovery |
B) Endurance / oxygen utilization
| Gene(s) | Explanation |
|---|---|
| ACE | endurance vs power tendency marker |
| HIF1A | oxygen response signaling |
| NOS3 | blood flow support |
| VEGF | capillary growth signaling |
| PPARGC1A / PPARA | aerobic capacity support pathways |
C) Power / sprint traits
| Gene(s) | Explanation |
|---|---|
| ACTN3 | fast-twitch muscle performance marker |
| CKM | muscle energy buffering |
| AMPD1 | ATP recycling during intense bursts |
D) Training response (adaptation rate)
| Gene(s) | Explanation |
|---|---|
| ACE / ACTN3 / PPARGC1A / NRF2 | adaptation, recovery, oxidative load handling |
| ACSL1 | fatty acid activation for energy use in muscle |
6) Nutrients pathways (absorption, activation, utilization)
A) Vitamin B12
| Gene(s) | Explanation |
|---|---|
| TCN2 | transport into cells |
| FUT2 | gut/secretor status; influences B12 availability via microbiome dynamics |
B) Choline
| Gene(s) | Explanation |
|---|---|
| PEMT / CHDH / BHMT | choline need & use in methylation/membranes |
C) Folate
| Gene(s) | Explanation |
|---|---|
| MTHFR / MTHFD1 | folate processing and availability |
D) Salt sensitivity
| Gene(s) | Explanation |
|---|---|
| ACE / AGT / AGTR1 / REN | RAAS control of fluid and pressure |
E) Vitamin D
| Gene(s) | Explanation |
|---|---|
| VDR | receptor response |
| GC | transport protein |
| CYP2R1 | activation step |
F) Caffeine
| Gene(s) | Explanation |
|---|---|
| CYP1A2 | primary caffeine metabolism speed |
| COMT | stimulant sensitivity via catechol handling |
G) Fatty acids
| Gene(s) | Explanation |
|---|---|
| FADS1 / FADS2 | omega-3/omega-6 conversion efficiency |
| ELOVL2 | long-chain fatty acid elongation |
| APOA2 / APOA5 / FABP2 | dietary fat response/handling |
H) Gluten
| Gene(s) | Explanation |
|---|---|
| HLA | celiac-associated immune recognition markers |
I) Iron overload
| Gene(s) | Explanation |
|---|---|
| HFE | iron absorption regulation |
J) Vitamin C
| Gene(s) | Explanation |
|---|---|
| SLC23A1 | vitamin C transport into cells |